While doing some deep learning neural network analysis for image recognition I ran across data that was provided in a file structure called HDF5. It was easy to use, and, as I discovered, the file system worked across many platforms, and was easily accessible using libraries and packages provided in both R and Python. This prompted me to do some research on the file system where I discovered that not only is it simple to use, can help in managing memory constraints (especially in R), and fast, it is portable across many platforms ranging from laptops running Windows 7, to OS X, to massively parallel processing (MPP) systems. Additionally, it is a free, open source platform for storing, archiving, and exchanging data. These are all good things, but I particularly like the last part, exchanging data. Anyway, rather than detailing all of the benefits of HDF5, I will direct you to their website where you can read more about it. In the meantime, here are some examples of how you use this file system in both R and Python.
Using deep learning neural networks for image recognition requires that the images be converted into vectors, and in a very particular order. Basically, the image comes in a variety of dimensions depending on the format, size, color, etc. For example, a jpeg image of 250px x 250px comes in three dimensions, each dimension corresponding to the intensity level of the RGB color spectrum. So a typical jpeg image in color will have the dimensions of 250x250x3. I won’t go into the details here because this process is detailed in another post: “Deep Learning & Neural Networks: Part 1“. If I wanted to share this data with someone, I can just send them the file. This way I don’t have to send them a directory full of images, and let them figure out how I randomly selected the images. All I have to do is send the file of processed data in an HDF5 file format. How do I go about doing that, and how can they use it?
Let’s assume I wrote my analysis in R, and I want to see if they get the same results using Python. The following code shows how I can load the image data, and save it in an HDF5 file format:
[snippet slug=createhdf5ofimages line_numbers=false lang=r]
The code above is simple and straightforward. In HDF5 you can create groups and within each group have multiple datasets. The way I chose to organize the data was by groups “traingroup”, “testgroup”, and “valgroup”, each group containing input feature variables, image file names, and the $$\hat y$$ values for each image (e.g., 1 = dog, 0 = no-dog). Once the data is saved in the HDF5 file format it is available for viewing using the HDFView gui as shown in the following figure:

Notice in the figure above that you can view the files to determine their content. The circled areas show where images were loaded in different formats. The front image consists of 750 vectors of length 187,500 (250 * 250 * 3), whereas the other file consists of 209 images with the dimensions 64x64x3. If the latter were formatted into vectors, like the former, the dimension would be 12288, 209 (64 * 64 * 3 = 12288).
Using the data from the dogvcat1.hr file, we will extract the data using both R and Python and display images:

[snippet slug=rreadhdf5file line_numbers=false lang=r]
Notice a couple of things from the code: the groups are loaded and data is now accessible from within each group. Datasets from the “traingroup” are extracted, and you now have access to your training data. The same is true of each group, however, notice that the size of the train_x group is only 1.8Kb, not the 1Gb that the variable is actually pointing to. This allows you to load the data as needed, delete it to make room for other data, and recall the data as needed.
With the data loaded, you can use it to extract data as needed without using up all your memory. The following shows how to extract data from a “traingroup” object, and reconstruct an image from the feature vector:
[snippet slug=rreconstituteimage line_numbers=false lang=r]
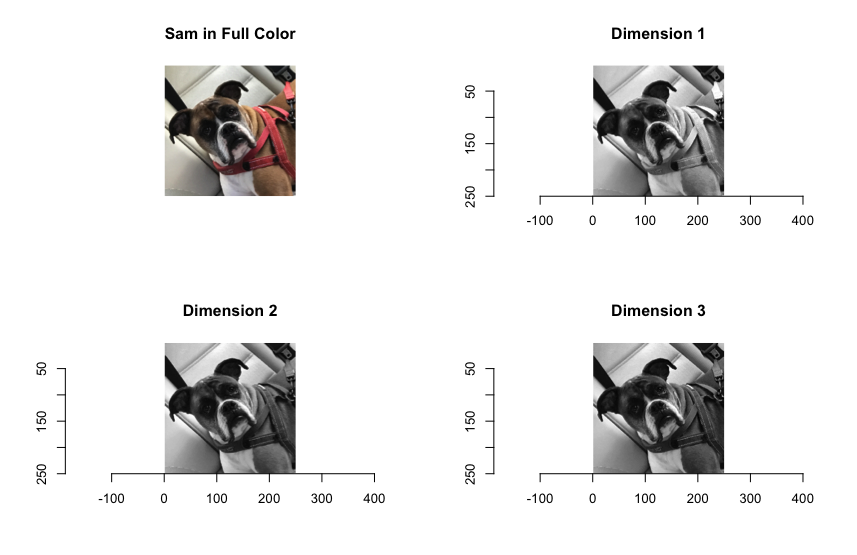
You will notice that the R library shows the various intensity levels for each color spectrum in black and white, while Python shows intensities with colors.
Accomplishing the same thing in Python 3:

[snippet slug=readhdf5filepython line_numbers=false lang=python]
With pointers to the data loaded, you can then access the data as needed. This particular dataset is too large to load all data at once for most computers, so we will load the training data only, select an image by index, and show multiple ways in Python that the image vector can be used to reconstitute the image object:
[snippet slug=pythonvectorintoimage line_numbers=false lang=python]

That’s all there is to it. The HDF5 file system is extremely easy to work with, it saves time, makes it easier to manage large datasets with relatively little memory, and for larger scale analysis, is capable of running on a MPP system.

